In this post, we will train a deep learning model. Plant disease recognition.
The use of deep learning and machine learning is helping solve many agricultural problems. A deep learning-based computer vision system was especially helpful. Such deep learning systems/models can easily recognize various plant diseases when trained on a suitable dataset. For example, training a deep learning model to Recognize different types of leaf diseases in rice.
Most of the time, it’s a matter of getting a large, well-defined dataset to solve such problems. Deep learning models need to be trained on massive amounts of data to recognize plant diseases. But that doesn’t mean you can’t train a simple deep learning model and see if such a model works.
In fact, this blog post uses about 1,300 images for training a deep learning model. When it comes to recognizing plant diseases, this may not seem like a big deal. However, as we will see later, it is a good starting point and provides a deep learning model that works quite well.

In this blog post, we start simple and gradually expand to broader and more difficult problems for agricultural applications. I will solve more practical applications like this in future posts.
Points covered in post
- We start by describing the plant disease recognition dataset we will be using.
- The training and coding section covers:
- A deep learning model used to recognize plant diseases.
- Enhancements to apply to the image and how they affect the result.
- After training is complete, test the model and visualize the class activation map.
Let’s move on.
Plant disease recognition dataset
use Kaggle Plant Disease Recognition Dataset Train a deep learning model in this post.
The dataset contains leaf images of a variety of plants, whether they are affected by disease or not.
This dataset contains a total of 1530 images and has three classes:
- health: Disease-free leaves.
- powdery: Leaves affected by powdery mildew. This is a type of fungal disease that affects plants based on the time of year.You can read more about the disease here.
- rust: Rust can affect a wide variety of plants. This is also a type of fungal disease.you may read more about it here.
Below are some of the images from the dataset and the different types of diseases they are classified into.

As you can see, it is very easy to identify powdery rust with the naked eye. Additionally, you can know how well a deep learning-based image classification model performs on your dataset.
The dataset is already split into 3 sets.
- training: The training set contains 1322 images.
- inspection: The validation set contains 60 images.
- test: The test set contains 150 images.
The dataset is fairly balanced across the three classes. This is suitable for training deep learning models.
Before proceeding, you can download the dataset from: here When running a training experiment on your local system.
directory structure
Below is the structure of all the directories and files used in this project.
.
├── input
│ ├── Test
│ │ └── Test
│ │ ├── Healthy
│ │ ├── Powdery
│ │ └── Rust
│ ├── Train
│ │ └── Train
│ │ ├── Healthy
│ │ ├── Powdery
│ │ └── Rust
│ └── Validation
│ └── Validation
│ ├── Healthy
│ ├── Powdery
│ └── Rust
├── notebooks
│ └── visualize_augmentations.ipynb
├── outputs
│ ├── cam_results [150 entries exceeds filelimit, not opening dir]
│ ├── test_results [150 entries exceeds filelimit, not opening dir]
│ ├── accuracy.png
│ ├── loss.png
│ └── model.pth
└── src
├── cam.py
├── datasets.py
├── model.py
├── test.py
├── train.py
└── utils.py
- In the directory structure above, the extracted data set in the
inputdirectory. Contains three separate subdirectories. Images for each class reside in their respective folders. - of
outputsThe directory contains results obtained from training and testing deep learning models. - of
notebooksThe directory contains a single Jupyter Notebook for visualizing augmented images. - At the end,
srcThe directory contains Python code files. Some of the important contents of these files are described in subsequent sections of this post.
You can also access all the code files and the trained model by downloading the zip file for this article. If you want to train the model yourself, you can download the dataset and arrange it according to the structure above. input directory.
PyTorch version
The code for this article is PyTorch version 1.12.1 and Torchvision 0.13.0Later versions (if available) will also work.
Plant Disease Recognition Using Deep Learning and PyTorch
This section covers important practical aspects of the training pipeline and dataset preparation.
We won’t go into detail about Python files here. important part only. All code files are still available as downloadable zip files.
Deep Learning Model (ResNet34)
For training, pre-trained ResNet34 Connect to the network and fine-tune with a plant disease dataset.
download code
Use a Torchvision model already pretrained on the ImageNet dataset. Preparing a ResNet34 model is very easy and only requires a few lines of code. The next block contains The entire code that goes into model.py File.
import torch.nn as nn
from torchvision import models
from torchvision.models import ResNet34_Weights
def build_model(pretrained=True, fine_tune=True, num_classes=10):
if pretrained:
print('[INFO]: Loading pre-trained weights')
model = models.resnet34(weights=ResNet34_Weights.DEFAULT)
else:
print('[INFO]: Not loading pre-trained weights')
model = models.resnet34(weights=None)
if fine_tune:
print('[INFO]: Fine-tuning all layers...')
for params in model.parameters():
params.requires_grad = True
elif not fine_tune:
print('[INFO]: Freezing hidden layers...')
for params in model.parameters():
params.requires_grad = False
# Change the final classification head.
model.fc = nn.Linear(in_features=512, out_features=num_classes)
return model
As you can see, we need to change the final fully connected layer. The number of classes was changed from 1000 (for ImageNet) to 3 (for the plant disease recognition dataset).
Experiments with other models were also carried out, but without success. The ResNet family of models works pretty well, and perhaps ResNet50 does even better.
Preparing the dataset for training
We come to the preparation part of the dataset. All code is datasets.py File.
Here we describe the hyperparameters and extensions used to prepare the dataset.
IMAGE_SIZE = 224 # Image size of resize when applying transforms. BATCH_SIZE = 32
Select an image size with a resolution of 224×224. Higher resolutions work fine, but require more GPU memory and longer training times. From experimentation, lower resolutions did not work well.
As we will see later, for the batch size, 32 gave pretty good results. If you are training on your local system and are running out of GPU memory (OOM), try reducing your batch size. However, a smaller batch size may require longer training to reach the same accuracy as a batch size of 32. This has to be experimented with.
It also uses some transformations and extensions. Augmentation reduces overfitting and allows for longer workouts. The following snippets show the transformation/extension of the training and validation sets.
# Training transforms
def get_train_transform(image_size):
train_transform = transforms.Compose([
transforms.Resize((image_size, image_size)),
transforms.RandomHorizontalFlip(p=0.5),
transforms.RandomRotation(35),
transforms.RandomAdjustSharpness(sharpness_factor=2, p=0.5),
transforms.ToTensor(),
transforms.Normalize(
mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225]
)
])
return train_transform
# Validation transforms
def get_valid_transform(image_size):
valid_transform = transforms.Compose([
transforms.Resize((image_size, image_size)),
transforms.ToTensor(),
transforms.Normalize(
mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225]
)
])
return valid_transform
For the training set, we choose three extensions. They are horizontal flip, random rotation, and random application of sharpness to the image.
For reference, here are some of the images augmented from the training set.

Otherwise, use ImageNet normalized values for both training and validation sets. This is because we are fine-tuning a pre-trained ResNet34 model.
The rest of the dataset preparation code involves creating the dataset and data loader.
Training a ResNet34 model for plant disease recognition
In this section you will run the training script. train.pyto see the results.
Before that, let’s take a look at the hyperparameters and parameters we use for training.
- Train for 20 epochs.You can change this by passing
--epochsArguments of the running command linetrain.py. - The optimizer is SGD with momentum and learning is 0.001. The learning rate can also be passed from the command line.use
--learning-ratediscussion on this. - Since this is a multiclass problem, we are using the Cross-Entropy loss function.
To start training, type the following command src directory.
python train.py --epochs 20
Below is a truncated portion of the output from the terminal.
[INFO]: Number of training images: 1322 [INFO]: Number of validation images: 60 [INFO]: Classes: ['Healthy', 'Powdery', 'Rust'] Computation device: cuda Learning rate: 0.001 Epochs to train for: 20 [INFO]: Loading pre-trained weights [INFO]: Fine-tuning all layers... 21,286,211 total parameters. 21,286,211 training parameters. [INFO]: Epoch 1 of 20 Training 100%|████████████████████████████████████████████████████████████████████| 42/42 [00:26<00:00, 1.57it/s] Validation 100%|██████████████████████████████████████████████████████████████████████| 2/2 [00:02<00:00, 1.24s/it] Training loss: 0.530, training acc: 77.912 Validation loss: 0.086, validation acc: 100.000 -------------------------------------------------- . . . [INFO]: Epoch 20 of 20 Training 100%|████████████████████████████████████████████████████████████████████| 42/42 [00:25<00:00, 1.63it/s] Validation 100%|██████████████████████████████████████████████████████████████████████| 2/2 [00:02<00:00, 1.19s/it] Training loss: 0.003, training acc: 99.924 Validation loss: 0.001, validation acc: 100.000 -------------------------------------------------- TRAINING COMPLETE
As you can see, we get almost 100% training accuracy. Validation accuracy is already 100%. Due to augmentation, training accuracy may be slightly lower. Training a few more epochs also seems to improve the training results slightly.
Let’s take a look at the Accuracy vs Loss graph.
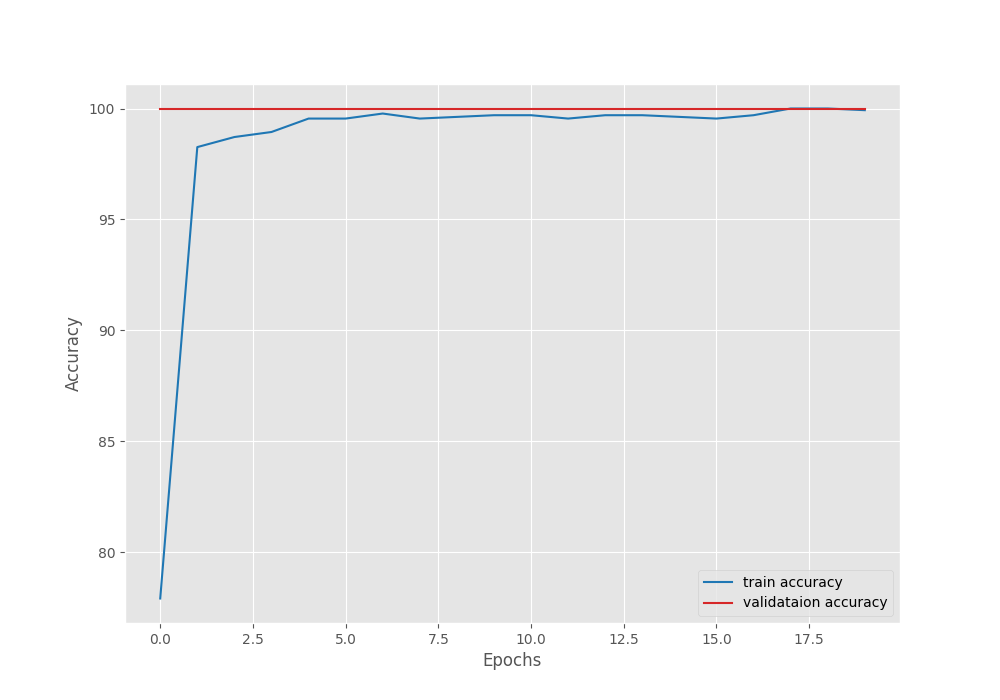
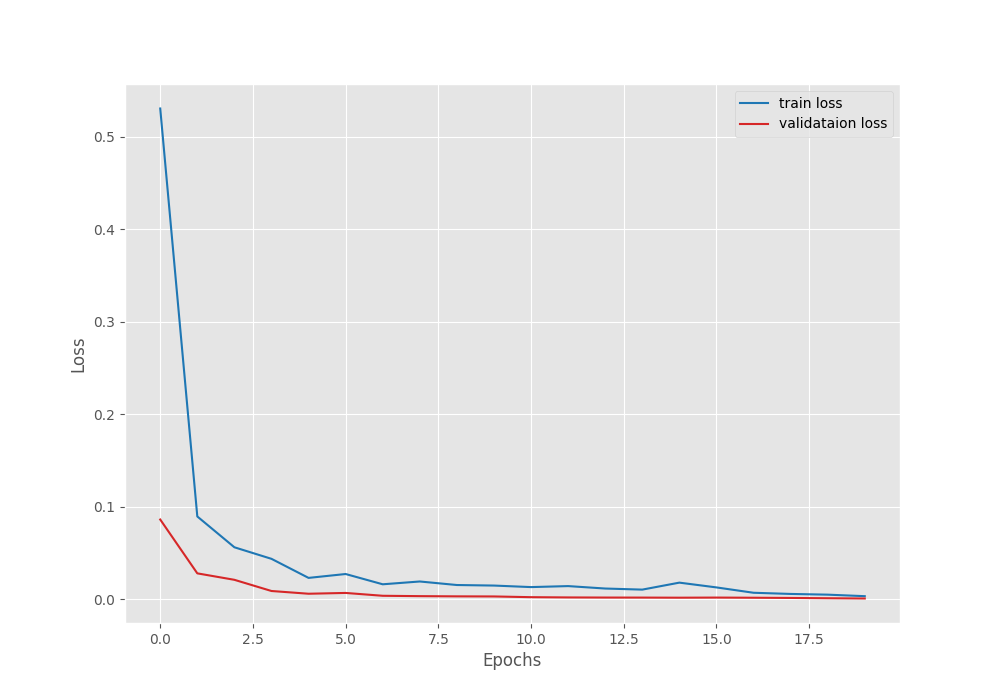
As you can see, the validation accuracy is already 100% from the first epoch. this is, momentum Specify coefficients with the SGD optimizer.Without momentum factor, convergence is slightly slower.
Test a trained model for plant disease recognition
A trained model is already available. This section does two things.
- to run the trained model on test images.
test.pyscript. - to visualize the class activation map on the test image.
cam.pyscript.
To run the test script, run the following command in your terminal:
python test.py
Below are the results.
python test.py [INFO]: Not loading pre-trained weights [INFO]: Freezing hidden layers... Testing model 100%|██████████████████████████████████████████████████████████████████| 150/150 [00:04<00:00, 33.20it/s] Test accuracy: 97.333%
The currently trained model gives over 97% accuracy.
Below are some of the results that are saved to disk.

The image above shows some of the correct predictions made by the model.

Figure 7 It shows two incorrect predictions made by the model.
For the image on the left, the ground truth is PowderyBut it looks like a labeling error and Rust disease. In the image on the right, the model predicts disease as: Powdery whereas in fact Rust.
Now to visualize the class activation map, run the following command:
python cam.py
The above also runs a test of the model. But what we are interested in is outputs/cam_results directory.

We can clearly see that the model focuses specifically on the leaf disease area when making predictions. It is very clear from Rust A disease where the model looks at spots and makes predictions.
Summary and conclusion
In this blog post, we used deep learning to solve a real-world problem on a small scale. Trained his ResNet34 model for recognizing plant diseases. After training the model, we tested it on the test set and also visualized the class activation map. This gave us a better insight into what the model was looking at when making predictions. I hope you found this post helpful.
If you have any questions, thoughts or suggestions, leave them in the comments section. They will definitely be addressed.
you can contact me using contact section.you can find me too LinkedInWhen twitter.

